Generation of hypoimmunogenic induced pluripotent stem cells by CRISPR-Cas9 system and detailed evaluation for clinical application
RESEARCHES
Key Points
- To reduce the risk of immune rejection, iPS cells were gene edited using CRISPR-Cas9 (1) techology to knock out the expression of three genes: HLA-A , HLA-B and CIITA
- Many iPS cells were found to have gen omic abnormalities following the gene editing, such as unexepected deletions and chromosomal translocations (2) , but iPS cells with these abnormalities could be identified and selected
- The CiRA Foundation is investigating these gene-edited iPS cells for clinical use
Summary
For iPS cells to be used in regenerative medicine, the avoidance of immune rejection is essential. One solution researchers are considering is the gene editing of human leukocyte antigen (HLA)(3) genes. In a new study published on June 11 in Molecular Therapy - Methods & Clinical Development, CiRA Foundation researchers report the simulatenous editing of three genes, HLA-A, HLA-B, and CIITA, in HLA homozygous(4) iPS cells by good manufacturing practice(5). The use of HLA homozygous iPS cells instead of HLA heterozygous iPS cells simplifies the gene editing process. Tests confirmed the iPS cells functioned as expected and could differentiate into cardiomyocytes. Whole genome sequencing detected several genetic and chromosomal abnormalities, such as copy number variations(6), but cells without these unwanted changes could be identified and selected. The production and quality control of these gene-edited HLA homozygous iPS cells provide a basis for their clinical use.
Scientific background
CRISPR-Cas9 genome editing technology was recently used to selectively knock out in iPS cells the expression of genes responsible for an immune response (see Xu and Wang et al.,Cell Stem Cell, 2019). These iPS cells, in which class I HLA-A and HLA-B and class II HLA genes (CIITA) were knocked out, were shown not to activate killer T cells(7) or NK cells(8), which are responsible for immune responses in the body. In that previous study, the knockout process took a significant amount of time. Shortening the timeframe may be possible by knocking out the genes simultaneously. However, this risks increasing the number of off-target mutations(9) and other genetic and chromosomal abnormalities in the iPS cells. This simulataneous gene editing is technically easier in iPS cells made from HLA homozygous donors, like those stocked at the CiRA Foundation, than HLA heterozygous donors, who are more common in the general population. At the same time, detailed assessment for the clinical use of iPS cells modified by this simultaneous gene editing approach has not been done.
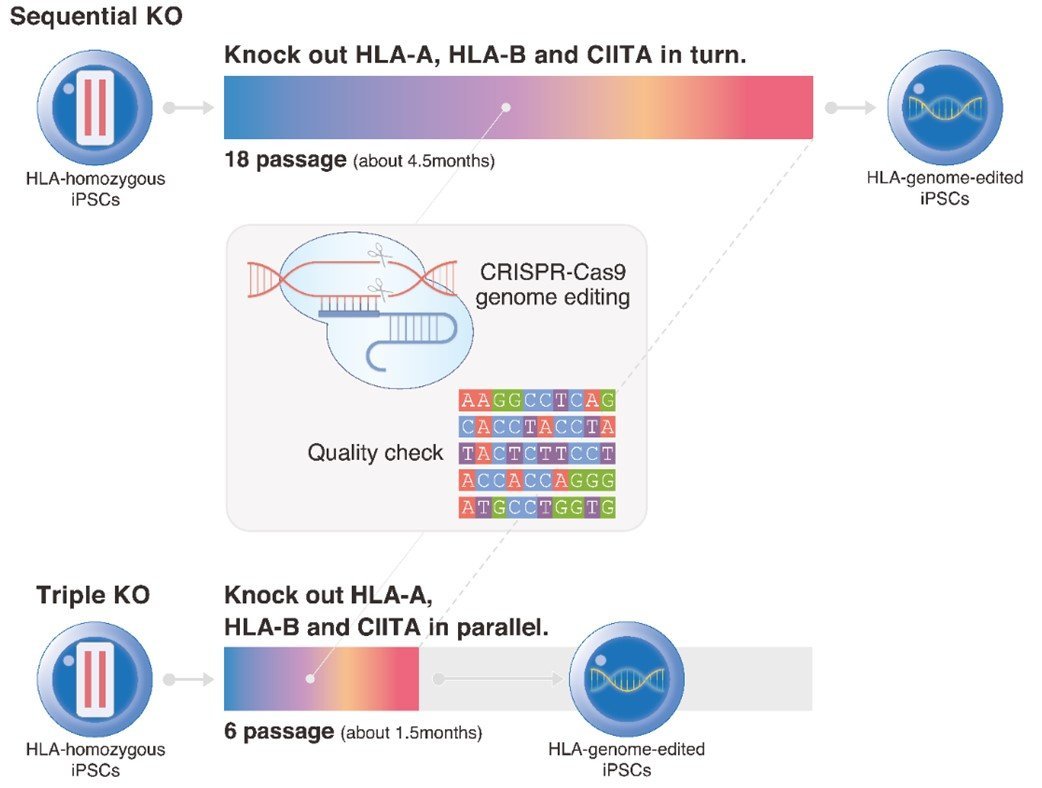
Schematic diagram of the gene editing timeframe. Knocking out HLA-A, HLA-B, and CIITA sequentially in iPS cells requires about 4.5 months, whereas knocking them out simultaneously takes only about 1.5 months and reduces the number of cell passages, which reduces the risk of unwanted mutations.
Main results
The researchers reported a method to simultaneously knock out the HLA-A, HLA-B, and CIITA genes in iPS cells (Fig. 1). In the CRISPR-Cas9 system, a Cas9 protein and gRNA are needed to edit a single gene. Using HLA homozygous iPS cells, the study shows how one gRNA is sufficient for the gene editing of HLA-A and HLA-B. Following the introduction of the CRISPR-Cas9 package into the iPS cells, single cell cloning(10) and detailed analysis were conducted. Several unwanted genetic and chromosomal changes were discovered, including copy number variations and chromosomal translocations (Fig. 2). Further analysis allowed the researchers to select cells that showed only the targeted gene editing and not these unwanted effects.
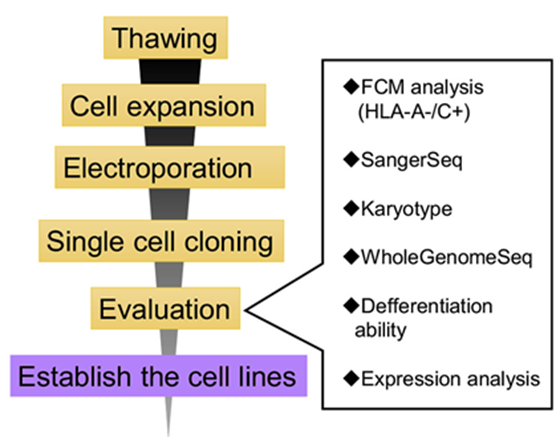
Figure1:A flowchart of the gene editing and quality assessment of the iPS cells.
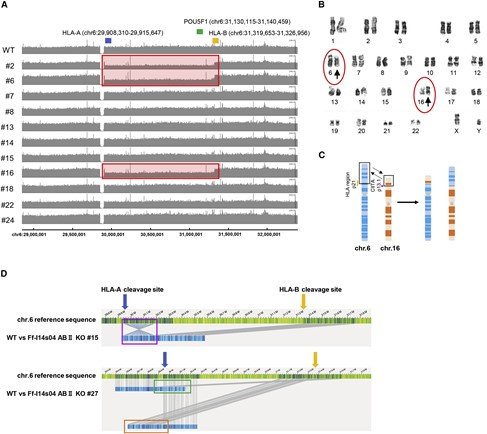
Figure2: Summary of genomic and karyotype analysis of iPS cells
A) Gene copy number analysis of the class I HLA gene cluster in chromosome 6. The positions of HLA A (blue), HLA B (orange), and POU5F1 (green) are shown. Of the 11 clones analyzed, 3 showed a decrease in the copy numbers of HLA A and HLA B (red boxes), indic ating large deletions.
B) K aryotyping results . Translocations were observed in chromosomes 6 and 16 (circled arrowheads).
C) A model for the translocation in chromosomes 6 and 16. The translocations were observed in 7 of the 30 iPS cell clones.
D) O ptial genome mapping revealed multiple chromosomal changes(blue bars).
Future work
This research has implications for the use of gene-edited iPS cells in clinical applications. It further demonstrates the importance of analysing in detail the effects of editing multiple genes simultaneously in HLA homozygous iPS cells. Based on the results of this study, the CiRA Foundation is developing such cells at its cell processing facilities for later provision to partner organizations.
Paper details
・Title
Generation of hypoimmunogenic induced pluripotent stem cells by CRISPR-Cas9 system and detailed evaluation for clinical application. doi.org/10.1016/j.omtm.2022.05.010
・Journal
Molecular Therapy - Methods & Clinical Development
・Authors
Yuko Kitano1, Sayaka Nishimura1, Tomoaki M Kato1, Anna Ueda1, Kaho Takigawa1, Masafumi Umekage1, Masaki Nomura1, Ayane Kawakami1, Haruna Ogawa1, Huaigeng Xu2,3, Akitsu Hotta2, Naoko Takasu1, Masayoshi Tsukahara1*
*corresponding author
・Author affiliations
1.The CiRA Foundation
2.The Center for iPS Cell Research and Application (CiRA), Kyoto University
3.Eli and Edythe Broad Center of Regeneration Medicine and Stem Cell Research, University of California San Francisco
Funding
Japan Agency for Medical Research and Development(AMED; Grant number: JP20bm0104001h0108)
Glossary
1) CRISPR-Cas9
Technology that cuts genomic DNA for gene editing. The founders of this technology were awarded the Nobel Prize in 2020.
2) Chromosome translocation
A break in chromosomes that results in the fragments relocating to different chromosomes.
3) Human leukocyte antigens (HLA)
A set of genes that code for proteins expressed on almost all cells in the body. They serve as a marker to the immune system to distinguish self cells from foreign cells.
4) HLA homozygous iPS cells
iPS cells in which both alleles (one from each parent) have the same HLA type. The CiRA Foundation has strategically prepared and distributes HLA homozygous iPS cells that HLA-match approximately 40% of the Japanese population, reducing the risk of immune rejection in this population.
5) Good manufacturing practice
Meets the minimal national requirements for the manufacturing of pharmaceuticals and other medical products.
6) Copy number variation
Refers to a gene that has an abnormal copy number.
7) Killer T cells
Immune cells that detect and respond to HLA Class I on other cells.
8) NK (natural killer) cells
Immune cells that detect and respond to a different set of cells than killer T cells.
9) Off-target mutations
Unwanted or undesired mutations that occur by the gene editing process.
10) Single cell cloning
The replication of a single cell from a cell population sto create a new cell population that is genetically identical.
Inquiries
For research and media inquiries
The CiRA Foundation
Public Relations Group
TEL:+81-80-2359-8495
E-mail:contact*cira-foundation.or.jp
(please change the * to @)
For AMED inquiries
Japan Agency for Medical Research and Development (AMED)
Department of Regenerative Medicine andCell and Gene Therapies
TEL:+81-3-6870-2220
E-mail:saisei*amed.go.jp
(please change the * to @)
Press release
